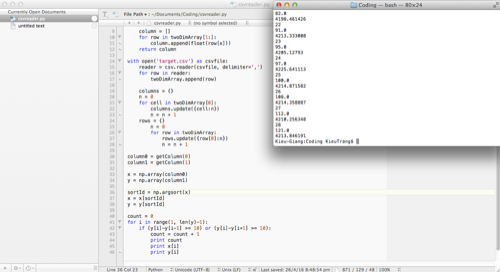
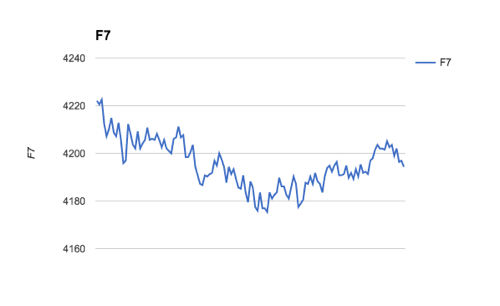
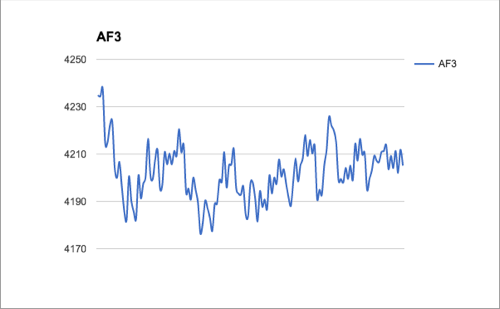
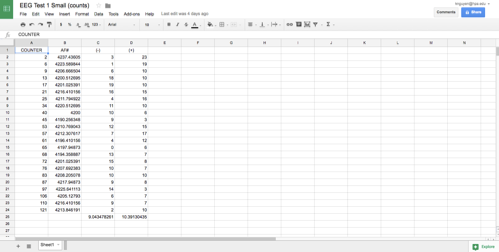
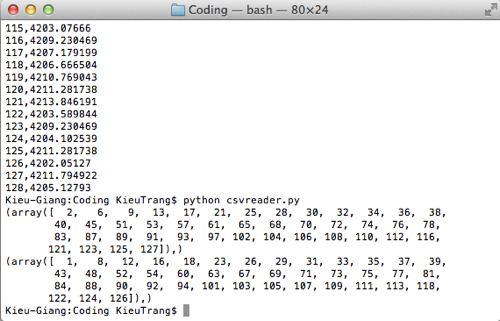
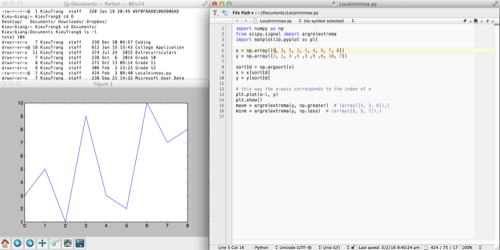
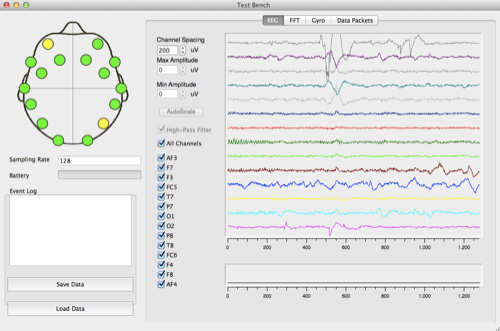
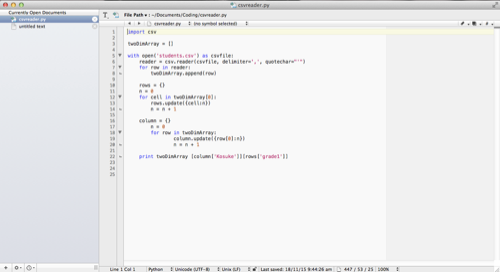
This week I made a lot of progress on the software development. I found a piece of Python code on StacksOverflow that help me find relative extremum:
import numpy as np
from scipy.signal import argrelextrema
import matplotlib.pyplot as plt
x = np.array([6, 3, 5, 2, 1, 4, 9, 7, 8])
y = np.array([2, 1, 3, 5, 3, 9, 8, 10, 7])
sortId = np.argsort(x)
x = x[sortId]
y = y[sortId]
# this way the x-axis corresponds to the index of x
plt.plot(x-1, y)
plt.show()
maxm = argrelextrema(y, np.greater) # (array([1, 3, 6]),)
minm = argrelextrema(y, np.less) # (array([2, 5, 7]),)
Here is the console when I try to run this code:
I already have my raw data demonstrated in lists and the code base to find the relative minimum/maximum, and now my task is to combine these two separate results together. I plan to continue working on this next week by developing and understanding the code as well as applying it to the real data.