Casting
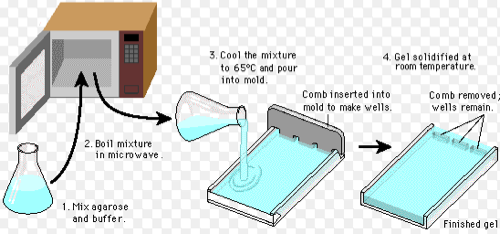
Today we cast a gel in order to run gel electrophoresis. We use .6g of agarose powder, 30ml of TBE buffer and 3ul of Sybr Safe DNA stain. The Sybr Safe intercalates between the base pairs in the double-stranded DNA and fluoresces green/yellow under blue light (thus, we use a blue light transilluminator). The agarose powder and TBE buffer are combined and then heated. Once the agarose has completely dissolved into the TBE, it is poured into the gel casting tray. The casting tray has a comb in it that creates well that we place the DNA in to run electrophoresis.
All in all, the entire process looks like this.
Hopefully we can run gel electrophoresis tomorrow...
All in all, the entire process looks like this.

Hopefully we can run gel electrophoresis tomorrow...


